Research grants in action


Brianna Maslen, a PhD student at Charles Sturt University, is working on a project focused on investigating the relationship between relative abundance of different microorganisms in rectal faecal samples taken from Angus calves. Brianna is being supported in her project by an Angus Foundation Research Grant, and by data generated through the Angus Sire Benchmarking Program.
Project Title: Evaluate whether microorganisms in rectal faecal samples can be profiled in order to identify their relationship with 1. Immune response 2. Growth performance 3. Meat Quality
The faecal microbial profiles are indicative of the distribution of microorganisms in the hind gut, which in turn have been shown to influence growth performance and immunity of beef cattle.
Brianna aims to investigate whether the faecal microbial profiles of cattle with high or low weight gain; or high or low immune responses, are significantly different.
She has collected and sequenced ~450 faecal samples from Angus weaners and an additional ~60 samples from yearling steers. Preliminary analyses indicates that there are indeed significant differences between faecal profiles of Angus yearling steers with high and low growth rates (See figure below).
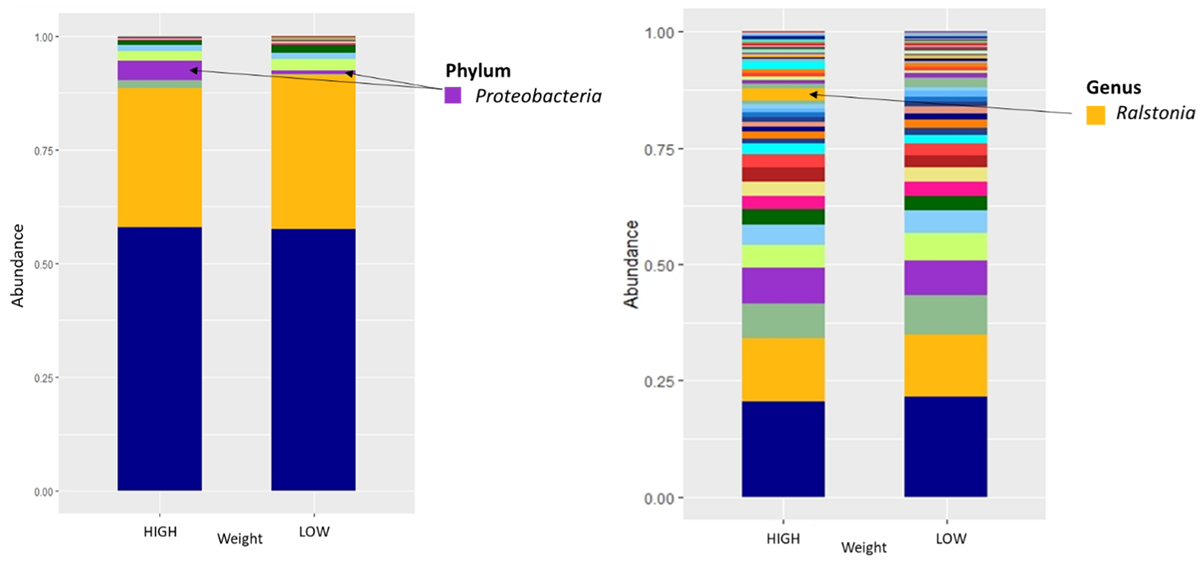

The figure presents the relative abundances (in percentages) of different phyla and genus (groups of microorganisms) in faecal samples from yearling steers with high or low growth rates. The colour of each bar represents a different phyla or genus, and each bar represents a different weight group.
A key preliminary observation relates to the relatively higher abundance of a microorganism called ‘Ralstonia’ that has been implicated with weight gain in studies focused
on obesity.
Once characterised, these findings could be used to identify underperforming cattle, and inform management strategies to improve performance of these cattle.
Further research is also ongoing, which is focused on investigating whether the relative abundance of different microorganisms in the hind gut is heritable, and if so, what genes/QTLs influence these abundances. This could afford opportunities for genetic selection of Angus cattle, so that they have desirable faecal profiles.
Feature Image: Brianna Maslen
By Brianna Maslen – Charles Sturt University, Wagga Wagga, NSW